Atlantic salmon from large Scottish east coast rivers - genetic stock identification: report
A report which investigates the potential to sample the genetic constitution of Atlantic salmon to work out which rivers they came from and whether it was possible to distinguish fish from among the large east coast rivers of Scotland.
Results
Quality Control
After the various QC measures outlined above were taken, combined with the full sib analysis, 125 individuals were removed, the final dataset contained genotypes for 69 microsatellites of 915 individuals from 40 sites, with an average N = 23 individuals per site. The average number of alleles per marker was 9.4 (± 4.2).
Genetic diversity
Details on within-site genetic diversity are given in Table 1, including average values across sites, together with the standard deviation (SD). After correction for multiple tests, genotypes in thirteen sites deviated significantly from the HW equilibrium.
| Site/s | Sites (NEPS or combined) | N | Ar | Ho | He | F | pHW |
|---|---|---|---|---|---|---|---|
| 85 | KSFT_Shin_5 | 26 | 3.33 | 0.49 | 0.50 | 0.04 | 0.070 |
| 86 | KSFT_Shin_7 | 29 | 3.54 | 0.53 | 0.52 | -0.02 | 0.008 |
| 87 | Kyle_Sutherland_2523 | 47 | 3.35 | 0.50 | 0.52 | 0.03 | < 0.0001 |
| 59 | Spey_001 | 46 | 3.88 | 0.52 | 0.55 | 0.07 | < 0.0001 |
| 60 | Spey_019 | 24 | 4.07 | 0.54 | 0.56 | 0.05 | < 0.0001 |
| 61 | Spey_024 | 29 | 3.71 | 0.53 | 0.54 | 0.03 | 0.013 |
| 62 | Spey_060 | 27 | 3.76 | 0.54 | 0.55 | 0.02 | 0.753 |
| 63 | Spey_138 | 29 | 3.87 | 0.52 | 0.54 | 0.05 | 0.009 |
| 64 | Spey_139 | 28 | 3.76 | 0.54 | 0.54 | 0.01 | 0.094 |
| 18 | Deveron_1623 | 15 | 3.53 | 0.53 | 0.52 | -0.02 | 0.992 |
| 19, 26 | Deveron_B | 17 | 4.17 | 0.54 | 0.55 | 0.01 | 0.334 |
| 20, 28, 29, 35 | Deveron_low | 22 | 3.99 | 0.52 | 0.55 | 0.10 | < 0.0001 |
| 31, 33, 34 | Deveron_mid | 18 | 3.77 | 0.55 | 0.54 | -0.02 | 0.585 |
| 22, 24, 27, 30, 32 | Deveron_upmost | 13 | 3.35 | 0.51 | 0.49 | -0.05 | 0.643 |
| 21, 25 | Deveron_upper | 24 | 3.87 | 0.53 | 0.54 | 0.02 | 0.417 |
| 37 | Don_1809 | 31 | 4.55 | 0.48 | 0.65 | 0.26 | < 0.0001 |
| 39 | Don_1827 | 26 | 3.95 | 0.54 | 0.55 | 0.03 | 0.010 |
| 40 | Don_1844 | 23 | 3.52 | 0.42 | 0.47 | 0.19 | < 0.0001 |
| 43 | Don_1934 | 11 | 3.12 | 0.43 | 0.37 | -0.08 | < 0.0001 |
| 38, 41 | Don_lower | 16 | 3.00 | 0.26 | 0.25 | -0.05 | 0.024 |
| 36, 42 | Don_middle | 16 | 3.51 | 0.39 | 0.45 | 0.20 | < 0.0001 |
| 11 | Dee_186 | 19 | 4.17 | 0.46 | 0.57 | 0.20 | < 0.0001 |
| 12 | Dee_190 | 47 | 3.93 | 0.52 | 0.54 | 0.07 | < 0.0001 |
| 13 | Dee_195 | 15 | 3.65 | 0.54 | 0.52 | -0.02 | 0.996 |
| 14 | Dee_313 | 39 | 3.7 | 0.50 | 0.53 | 0.07 | < 0.0001 |
| 15 | Dee_314 | 29 | 4.65 | 0.48 | 0.63 | 0.25 | < 0.0001 |
| 16 | Dee_316 | 29 | 3.71 | 0.53 | 0.53 | 0.04 | 0.014 |
| 17 | Dee_345 | 14 | 5.27 | 0.33 | 0.27 | -0.19 | 0.073 |
| 48 | Esk_2000 | 28 | 3.79 | 0.54 | 0.53 | 0.00 | 0.334 |
| 51 | Esk_2027 | 10 | 3.68 | 0.53 | 0.52 | -0.04 | 0.801 |
| 56 | Esk_2111 | 15 | 4.32 | 0.52 | 0.60 | 0.14 | < 0.0001 |
| 47, 50, 52 | Esk_Luther | 16 | 4.07 | 0.51 | 0.55 | 0.07 | 0.003 |
| 68 | Tay_4106 | 24 | 3.63 | 0.55 | 0.53 | -0.04 | 0.003 |
| 72 | Tweed_4148 | 21 | 3.71 | 0.52 | 0.52 | 0.01 | 0.295 |
| 73 | Tweed_4153 | 21 | 3.43 | 0.50 | 0.51 | 0.04 | 0.036 |
| 76 | Tweed_4159 | 12 | 3.34 | 0.51 | 0.48 | -0.06 | 0.997 |
| 77 | Tweed_4160 | 14 | 3.69 | 0.53 | 0.53 | 0.00 | 0.118 |
| 79 | Tweed_4276 | 16 | 3.7 | 0.53 | 0.52 | -0.01 | 0.797 |
| 71, 74 | Tweed_Leader | 16 | 3.65 | 0.50 | 0.51 | 0.05 | 0.148 |
| 70, 75 | Tweed_upper | 13 | 3.65 | 0.52 | 0.51 | -0.01 | 0.826 |
| Mean | 23 | 3.78 | 0.50 | 0.52 | 0.04 | ||
| SD | 10 | 0.42 | 0.06 | 0.07 | 0.09 |
Genetic structure
Supplementary table S1 details the differentiation parameters FST and DA. Average and SD values for those were 0.096 ± 0.122 and 0.317 ± 0.465, respectively. 507 out of the 780 (65%) pairwise comparisons were significant after FDR correction.
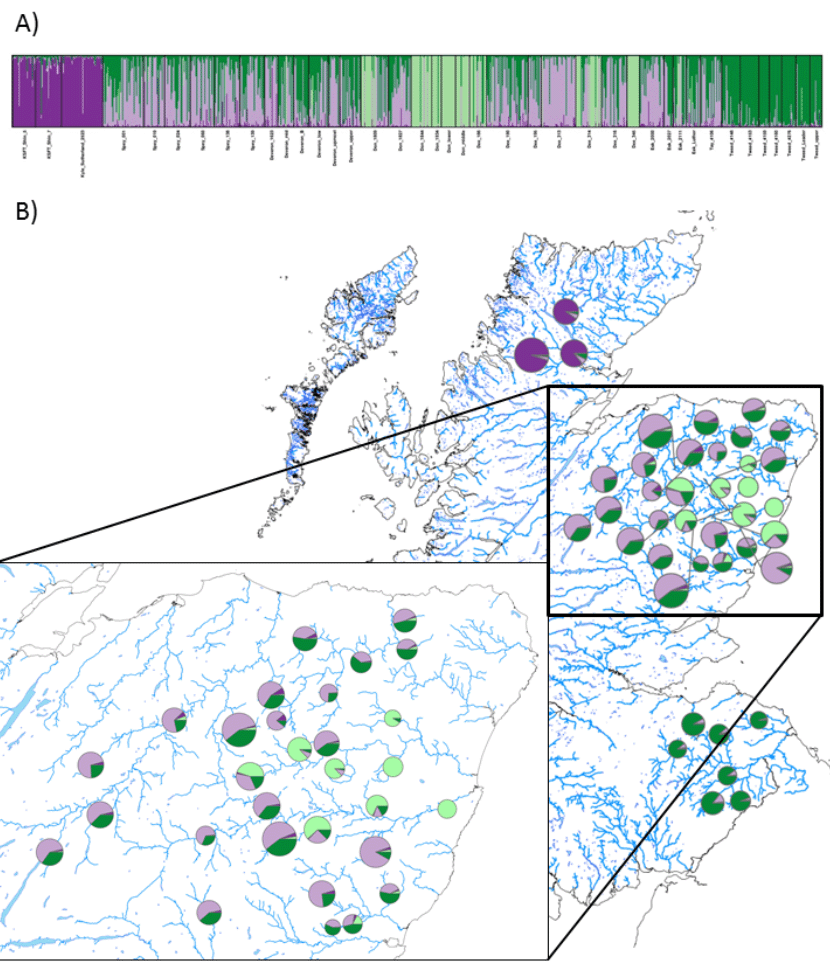
The optimal number of clusters in the dataset was K = 4. At that level, the sites from the rivers Oykel/Cassley/Shin and those from the River Tweed each form their own cluster (1: purple and 2: dark green in Fig. 2A, respectively). The majority of the sites from the River Don and a few from the River Dee (Dee_186, Dee_314 and Dee_345) are grouped in a third cluster (light green in Fig. 2A). The remainder of the sites encompassed the fourth group (lila in Fig. 2A). A geographical presentation of the clustering results is also given in Figure 2B.
Self-assignment
At the first river-level and based on all fish (i.e. not using a probability cut-off), assignment accuracy varied from 33.3% for the River North Esk to 94.1% for the River Oykel (Table 2). After applying the 0.8 probability cut-off level, assignment accuracies increased. However, only the rivers Oykel/Cassley/Shin and Tweed met the required assignment accuracy condition (80% accuracy). These two rivers were thus retained as separate assignment units. The misassignments, together with geographic proximity, were used to combine low accuracy rivers into larger assignment units. This was carried out by combining the adjacent rivers Spey and Deveron into a new unit (SpeyDev); and the Don and Dee into another new unit (DonDee). Self-assignment was then performed to the new Level 2 assignment units, using the 0.8 probability cut-off.
Results of the first, river-level, assignment analysis. Table 2 presents the results of all fish assignments, whilst Table 3 includes the results when a probability cut-off of 0.8 is applied. Both accuracy and number of fish assigned for each river is shown in bold.
| All assignments | Assigned to | |||||||
|---|---|---|---|---|---|---|---|---|
| River of origin | Oykel/Cassley/Shin | Spey | Deveron | Don | Dee | North Esk | Tay | Tweed |
| Oykel/Cassley/Shin | 111 | 2 | 1 | 1 | 0 | 2 | 0 | 1 |
| Spey | 1 | 132 | 12 | 14 | 21 | 16 | 2 | 4 |
| Deveron | 1 | 41 | 29 | 8 | 19 | 10 | 1 | 4 |
| Don | 0 | 20 | 3 | 98 | 9 | 1 | 3 | 6 |
| Dee | 3 | 52 | 8 | 63 | 75 | 19 | 2 | 7 |
| North Esk | 2 | 25 | 3 | 12 | 12 | 26 | 2 | 5 |
| Tay | 0 | 6 | 0 | 1 | 1 | 1 | 16 | 0 |
| Tweed | 0 | 14 | 6 | 11 | 7 | 3 | 1 | 84 |
| Accuracy % | 94.1 | 45.2 | 46.8 | 47.1 | 52.1 | 33.3 | 59.3 | 75.7 |
| Prob cut-off >0.8 | Assigned to | |||||||
|---|---|---|---|---|---|---|---|---|
| River of origin | Oykel/Cassley/Shin | Spey | Deveron | Don | Dee | North Esk | Tay | Tweed |
| Oykel/Cassley/Shin | 107 | 2 | 1 | 0 | 0 | 1 | 0 | 1 |
| Spey | 1 | 115 | 6 | 6 | 13 | 11 | 2 | 2 |
| Deveron | 1 | 29 | 28 | 4 | 10 | 6 | 1 | 3 |
| Don | 0 | 12 | 1 | 96 | 3 | 0 | 1 | 5 |
| Dee | 2 | 36 | 6 | 56 | 60 | 14 | 1 | 4 |
| North Esk | 0 | 20 | 3 | 10 | 8 | 24 | 1 | 4 |
| Tay | 0 | 6 | 0 | 1 | 0 | 1 | 16 | 0 |
| Tweed | 0 | 11 | 3 | 5 | 5 | 3 | 1 | 79 |
| Assigned % | 94.1 | 79.1 | 77.4 | 85.6 | 68.8 | 76.9 | 85.2 | 88.3 |
| Accuracy % | 96.4 | 49.8 | 58.3 | 53.9 | 60.6 | 40.0 | 69.6 | 80.6 |
At Level 2, both accuracy and percentage of fish assigned for SpeyDev and DonDee had increased, but were still not meeting the conditions required (Table 4). The majority of misassignments were between the regions SpeyDev, DonDee and the River North Esk. As such, these were combined into a larger unit, termed “East” and a Level 3 self-assignment carried out.
| Prob cut-off >0.8 | Assigned to | |||||
|---|---|---|---|---|---|---|
| Assignment unit | Oykel/Cassley/Shin | SpeyDev | DonDee | North Esk | Tay | Tweed |
| Oykel/Cassley/Shin | 107 | 3 | 0 | 1 | 0 | 1 |
| SpeyDev | 2 | 192 | 39 | 17 | 3 | 5 |
| DonDee | 2 | 59 | 222 | 14 | 2 | 9 |
| North Esk | 0 | 24 | 18 | 24 | 1 | 4 |
| Tay | 0 | 6 | 1 | 1 | 16 | 0 |
| Tweed | 0 | 14 | 12 | 3 | 1 | 79 |
| Assigned % | 94.1 | 84.7 | 82.5 | 76.9 | 85.2 | 88.3 |
| Accuracy % | 96.4 | 64.4 | 76.0 | 40.0 | 69.6 | 80.6 |
At Level 3, assignment accuracy of fish assigned to the “East” met the accuracy threshold (Table 5). However, the assignment accuracy of the River Tay was still below the set threshold and misassignments were to the “East” group. As such, the Tay site was included within the larger “East” region and a final self-assignment carried out.
| Prob cut-off >0.8 | Assigned to | |||
|---|---|---|---|---|
| Assignment unit | Oykel/Cassley/Shin | East1 | Tay | Tweed |
| Oykel/Cassley/Shin | 107 | 6 | 0 | 1 |
| East1 | 4 | 707 | 6 | 18 |
| Tay | 0 | 9 | 16 | 0 |
| Tweed | 0 | 32 | 1 | 79 |
| Assigned % | 94.1 | 95.7 | 88.5 | 90.7 |
| Accuracy % | 96.4 | 93.8 | 69.6 | 80.6 |
At the final level, all rivers/regions met the assignment accuracy threshold, with 93.8% of fish assigned with 95.9% accuracy to the three defined assignment units. Assignment details are given in Table 6.
| Prob cut-off >0.8 | Assigned to | ||
|---|---|---|---|
| Assignment unit | Oykel/Cassley/Shin | East2 | Tweed |
| Oykel/Cassley/Shin | 107 | 6 | 1 |
| East2 | 4 | 749 | 18 |
| Tweed | 0 | 33 | 79 |
| Assigned % | 94.1 | 96.8 | 90.7 |
| Accuracy % | 96.4 | 95.1 | 80.6 |
Contact
Email: ScotMER@gov.scot
There is a problem
Thanks for your feedback