Juvenile Atlantic salmon - mapping the early running component: report
This Scottish Marine and Freshwater Science report describes the application of a genetic tool to estimate the early running component of juvenile Atlantic salmon. The proportions were mapped to visualise within and between-river patterns. These maps can be used to inform and help river managers achieve their conservation aims.
Materials and Methods
Sample Collection
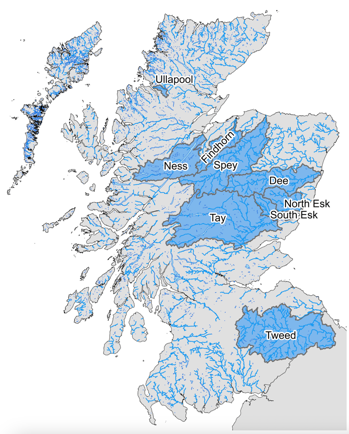
From 2017 to 2022, electrofishing was carried out in 231 sites across nine rivers: Ullapool, Ness, Findhorn, Spey, Dee, North Esk, South Esk, Tay and Tweed (Fig. 1, Table 1). Site selection was such as to ensure an as even a distribution throughout the salmon catchment area as was possible. A pelvic fin clip was taken from up to thirty anaesthetized juveniles. From each juvenile, it was noted whether it was a fry or parr, fork length was recorded and a scale sample was taken (with the exception of the smallest fry). The fish were then given time to recover before being released back into the river. Fin clips were stored in 99% ethanol.
River |
N sites |
N samples |
|---|---|---|
Dee |
42 |
1087 |
Findhorn |
17 |
486 |
Ness |
18 |
468 |
North Esk |
24 |
626 |
South Esk |
14 |
297 |
Spey |
45 |
1201 |
Tay |
35 |
864 |
Tweed |
27 |
744 |
Ullapool |
9 |
175 |
Genetic screening
The rationale behind the selection of the run timing panel, consisting of twelve SNPs, can be found in Cauwelier et al. (2024), whilst primer details are described in Cauwelier et al. (2018).
DNA from the fin clips was extracted using a Chelex extraction protocol, as described in Walsh et al. (1991). Extracts were processed for and screened on the Standard Biotools EP1 platform (Standard Biotools, San Francisco, CA, USA), following the manufacturer's protocols. This included an initial specific target amplification step, whereby all twelve SNPs were amplified in a multiplex, followed by a screening based on FlexSix or 192.24 integrated fluidic circuits (IFCs). Initial genotyping, performed in Standard Biotools genotyping software, was cross checked by a second investigator before data finalisation.
Data analysis
Genotyping scoring percentage was calculated for each individual. Poor-performing individuals (< 70%) were rescreened. Due to the rationale behind the SNP selection, whereby the major peaks were covered by two SNPs (see Cauwelier et al., 2024) to provide back-up in event of non-amplification, scoring percentage cut-off was set to 67% (8 out of the 12 SNPs). Samples not reaching the threshold value were removed from further analysis.
The baseline dataset, consisting of adult fish, as described in Cauwelier et al. (2018), were used as the reference individuals, with the different individual river and run timing groups (early v late) retained as separate assignment units. However, early and late run timing groups were combined across rivers to create two reporting groups. Individual assignment of each juvenile to the baseline run timing reporting groups was then carried out in ONCOR (Kalinowski et al. 2007), with probabilities estimated using the method of Rannala and Mountain (1997).
From the individual probabilities associated with the validation fish used in Cauwelier et al. (2018), the mean probability of the early running grilse (Apr-Jun) and MSW (Feb – Apr) was calculated, which was 0.828. This value was used as a cut off to assign fish as early running and was used to determine the early running component for each site by applying it to the individual probabilities. The proportion of early running fish was calculated by dividing the number of fish with a probability > 0.828 by the total number of fish for each site. This was then converted into a proportion, from which the proportion of the late returning component was derived (100 – proportion early). For each river, those proportions were visualised on a map using ArcGis (ArcGIS [ArcMap]. Version 10.6. Redlands, CA: Environmental Systems Research Institute, Inc., 2010)
Contact
Email: Eef.Cauwelier@gov.scot
There is a problem
Thanks for your feedback